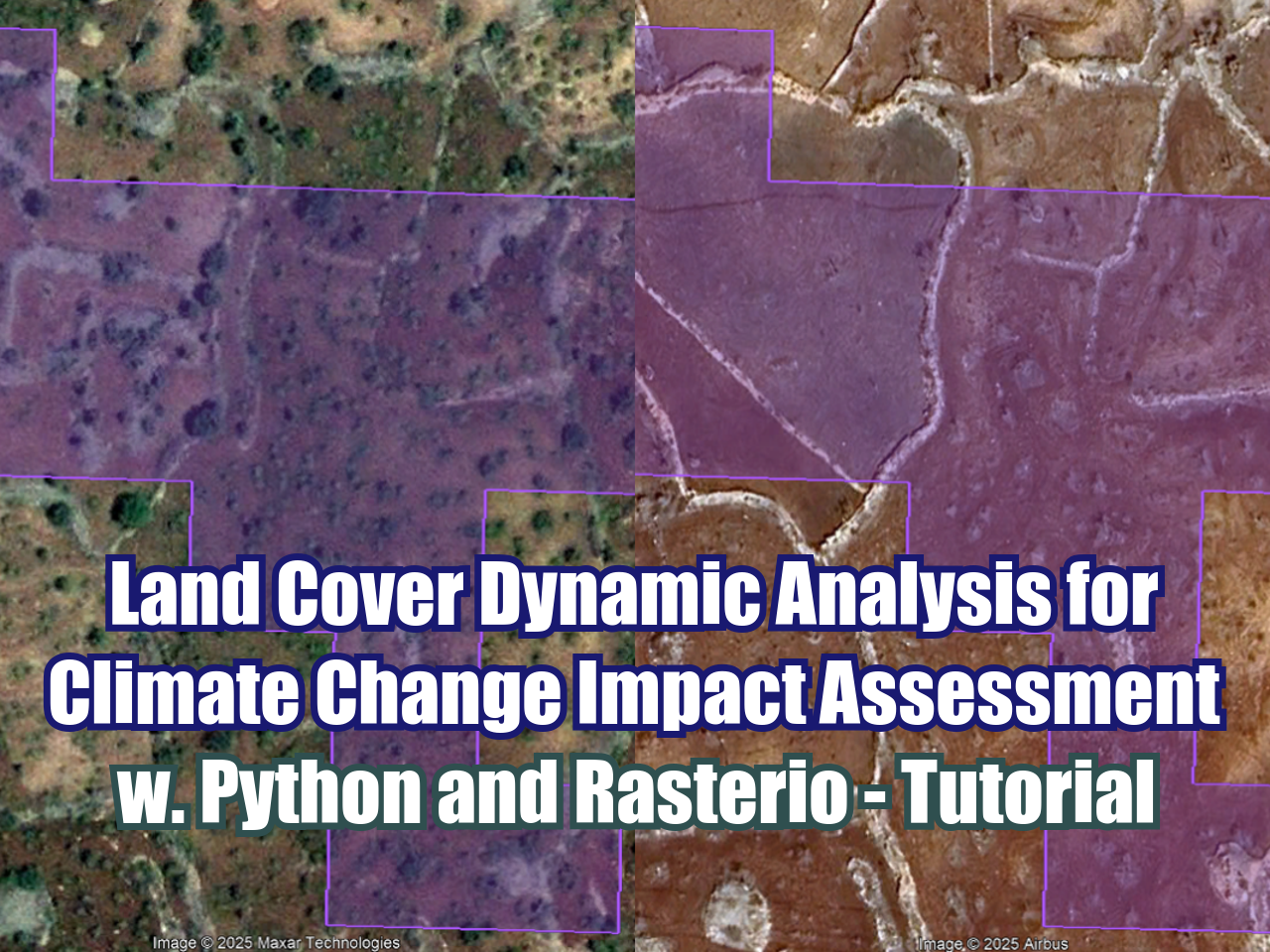
Land Cover Dynamic Analysis for Climate Change Impact Assessment with Python and Rasterio - Tutorial
/Satellite imagery brought us the capacity to see the land surface on recent years but we haven’t been so successful to understand land cover dynamics and the interaction with economical, sociological and political factors. Some deficiencies for this analysis were found on the use of GIS commercial software, but there are other limitations in the way we apply logical and mathematical processes to a set of satellite imagery. Working with geospatial data on Python gives us the capability to filter, calculate, clip, loop, and export raster or vector datasets with an efficient use of the computational power providing a bigger scope on data analysis.
This is an applied of land cover dynamic analysis over some regions of Turkey for the evaluation of land aridification and land abandonment due to climate change on the last decade. The case covers all the steps in Python with related geospatial libraries to process Sentinel 2 images for an area of study with considerations of hydrological conditions and reduction of outlier values.
Tutorial
Input data
https://owncloud.hatarilabs.com/s/N2rZop3wc60TEGL
Password to download data: Hatarilabs.
Code
Part 1
import os
import rasterio
from rasterio import plot
from rasterio.crs import CRS
import matplotlib.pyplot as plt
import numpy as np
from rasterio.mask import mask
from rasterio.warp import calculate_default_transform, reproject, Resampling
import geopandas as gpd
from affine import AffineaoiPath = '../shp/dagecitAreaBoundWgs37N.shp'
ndviDateList = [x for x in os.listdir('../rst') if '-' in x]
ndviDateListdef createTotalNdvi(ndviDate):
totalPath = os.path.join('../rst',ndviDate)
band4List = [x for x in os.listdir(totalPath) if x.endswith('B04.tif')]
band5List = [x for x in os.listdir(totalPath) if x.endswith('B05.tif')]
band4 = rasterio.open(os.path.join(totalPath,band4List[0]))
band5 = rasterio.open(os.path.join(totalPath,band5List[0]))
#generate nir and red objects as arrays in float64 format
red = band4.read(1).astype('float64')
nir = band5.read(1).astype('float64')
#ndvi calculation, empty cells or nodata cells are reported as 0
ndvi=np.where(
(nir+red)==0.,
0,
(nir-red)/(nir+red))
#export ndvi image
ndviImage = rasterio.open('../rst/ndviTotal/%s.tif'%ndviDate,'w',driver='Gtiff',
width=band4.width,
height = band4.height,
count=1, crs=band4.crs,
transform=band4.transform,
dtype='float32')
ndviImage.write(ndvi,1)
ndviImage.close()for ndviDate in ndviDateList:
createTotalNdvi(ndviDate)def reprojectRaster(ndvi):
inNdvi = os.path.join('../rst','ndviTotal',ndvi+'.tif')
outNdvi = os.path.join('../rst','ndviReproj',ndvi+'.tif')
with rasterio.open(inNdvi) as src:
dst_crs = "EPSG:32637"
transform, width, height = calculate_default_transform(
src.crs, dst_crs, src.width, src.height, *src.bounds)
kwargs = src.meta.copy()
kwargs.update({
"crs": dst_crs,
"transform": transform,
"width": width,
"height": height
})
with rasterio.open(outNdvi, "w", **kwargs) as dst:
for i in range(1, src.count + 1):
reproject(
source=rasterio.band(src, i),
destination=rasterio.band(dst, i),
src_transform=src.transform,
src_crs=src.crs,
dst_transform=transform,
dst_crs=dst_crs,
resampling=Resampling.nearest # or bilinear, cubic, etc.
)for ndviDate in ndviDateList:
reprojectRaster(ndviDate)outRes = 30
boundDf = gpd.read_file(aoiPath)
xDim = boundDf.total_bounds[2] - boundDf.total_bounds[0]
yDim = boundDf.total_bounds[3] - boundDf.total_bounds[1]
print(xDim,yDim)
ncol = int(xDim // outRes)
nrow = int(yDim // outRes)
print(ncol,nrow)print(boundDf.total_bounds)def clipRasterByMask(ndvi,boundDf):
inNdvi = os.path.join('../rst','ndviReproj',ndvi+'.tif')
outNdvi = os.path.join('../rst','ndviClip',ndvi+'.tif')
# Open the raster and apply the mask
with rasterio.open(inNdvi) as src:
out_image, out_transform = mask(src, boundDf.geometry.values, crop=True)
out_meta = src.meta.copy()
out_meta.update({
"driver": "GTiff",
"height": out_image.shape[1],
"width": out_image.shape[2],
"transform": out_transform
})
# Write the clipped raster
with rasterio.open(outNdvi, "w", **out_meta) as dest:
dest.write(out_image)for ndviDate in ndviDateList:
clipRasterByMask(ndviDate, boundDf)def resampleRasterByParams(ndvi,boundDf, ncol,nrow,outRes):
inNdvi = os.path.join('../rst','ndviClip',ndvi+'.tif')
outNdvi = os.path.join('../rst','ndviClipResampled',ndvi+'.tif')
with rasterio.open(inNdvi) as src:
transform = Affine(
outRes, 0, boundDf.total_bounds[0],
0, -outRes, boundDf.total_bounds[3]
)
kwargs = src.meta.copy()
kwargs.update({
'height': nrow,
'width': ncol,
'transform': transform
})
with rasterio.open(outNdvi, 'w', **kwargs) as dst:
reproject(
source=rasterio.band(src, 1),
destination=rasterio.band(dst, 1),
src_transform=src.transform,
src_crs=src.crs,
dst_transform=transform,
dst_crs=src.crs,
resampling=Resampling.bilinear
)for ndviDate in ndviDateList:
resampleRasterByParams(ndviDate,boundDf,ncol,nrow,outRes)Part 2
import os
import rasterio
from rasterio import plot
from rasterio.crs import CRS
import matplotlib.pyplot as plt
import numpy as np
from rasterio.mask import mask
from rasterio.warp import calculate_default_transform, reproject, Resampling
import fionandviDateList = [x for x in os.listdir('../rst/ndviReproj') if x.endswith('.tif')]
ndviDateListdef calculateMeanNdvi(raster1, raster2, rasterName):
raster1Path = os.path.join('../rst/ndviClipResampled',raster1)
raster2Path = os.path.join('../rst/ndviClipResampled',raster2)
rst1 = rasterio.open(raster1Path)
rst2 = rasterio.open(raster2Path)
rst1Band = rst1.read(1).astype('float64')
rst2Band = rst2.read(1).astype('float64')
meanNdvi=np.where(
(rst1Band+rst2Band)==0.,
0,
(rst1Band+rst2Band)/2)
ndviImage = rasterio.open('../rst/ndviMean/%s.tif'%rasterName,'w',driver='Gtiff',
width=rst1.width,
height=rst1.height,
count=1, crs=rst1.crs,
transform=rst1.transform,
dtype='float32')
ndviImage.write(meanNdvi,1)
ndviImage.close()calculateMeanNdvi(ndviDateList[0], ndviDateList[1], 'ndvi_2013_2014')calculateMeanNdvi(ndviDateList[2], ndviDateList[3], 'ndvi_2023_2024')def calculateNdviDifference(raster1,raster2,rasterName,threshold):
raster1Path = os.path.join('../rst/ndviMean',raster1)
raster2Path = os.path.join('../rst/ndviMean',raster2)
rst1 = rasterio.open(raster1Path)
rst2 = rasterio.open(raster2Path)
rst1Band = rst1.read(1).astype('float64')
rst2Band = rst2.read(1).astype('float64')
diffNdvi=np.where(
(rst1Band-rst2Band)>=threshold,
rst1Band-rst2Band,
-9999)
ndviImage = rasterio.open('../rst/ndviMean/%s.tif'%rasterName,'w',driver='Gtiff',
width=rst1.width,
height=rst1.height,
count=1, crs=rst1.crs,
transform=rst1.transform,
dtype='float32',
nodata=-9999)
ndviImage.write(diffNdvi,1)
ndviImage.close()calculateNdviDifference('ndvi_2013_2014.tif','ndvi_2023_2024.tif','dif_2013_2023',0.03)