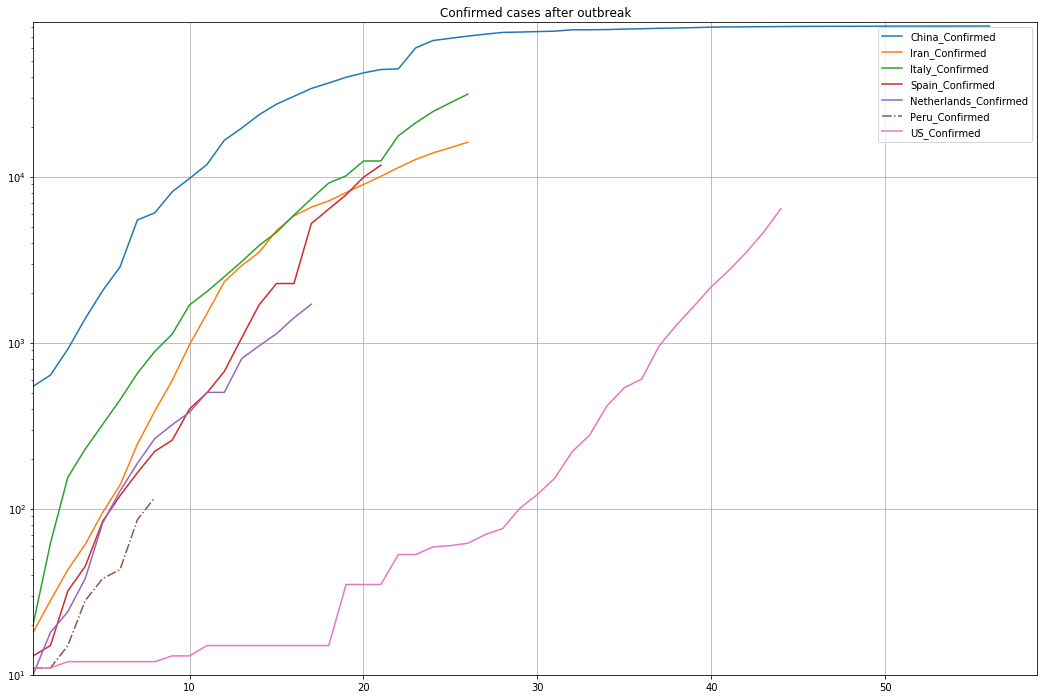
Decisions need to be made based on data, and the impact of those decisions (or the lack of them) are also analyzed on data. Nowadays we face a global pandemic of Coronavirus that has changed our normal lifestyles with confirmed cases and related deaths all over the world.
The temporal and spatial distribution of the Coronavirus cases need to be evaluated on a interactive platform for a enhanced data filtering, classification, plotting and comparative analysis. We have developed a webinar for the trend analysis of Coronavirus cases done with Python and Pandas on a Jupyter Network.
The webinar is based on the data from:
2019 Novel Coronavirus COVID-19 (2019-nCoV) Data Repository by Johns Hopkins CSSE ( https://github.com/CSSEGISandData/COVID-19 )
Content
The webinar cover the following topics:
Data scraper for the data repository
Compilation of daily data and name fixing
Plot confirmed/deaths/recovered cases by region
Plot confirmed/deaths/recovered cases by country
Group data by country and date
Generation of a “days after outbreak” attribute
Definition of plotting functions
Comparison of confirmed/deaths/recovered cases among countries
Instructor
Saul Montoya M.Sc.
Hydrogeologist - Numerical Modeler
Know more about his references here.
Instructions
For participants with computer in Windows, they need to have Anaconda installed from https://www.anaconda.com/distribution/#download-section on the Python3.7 option. Linux and Mac users have to install Jupyter through Pip.
Download the input data from this link, unzip and place under your “My Documents”
Download the Coronavirus database from this link, unzip and place under “My Documents/coronavirusTrendsandDistributionAnalysisPythonPandas”
The webinars will be held trough Youtube streaming. Links will be provided to your email.
Date and time
March 21, 2020 at 3 pm. Lima Time (GMT -5)
Estimated duration: 2h 15m.
Registration will be available till March 20, 2020 at 9 pm.